前面,我们生信技能树的讲师小洁老师与萌老师新开了一个学习班:《掌握Python,解锁单细胞数据的无限可能》,身为技能树的一员,近水楼台先得月,学起!下面是我的学习笔记,希望可以给你带来一点参考
今天继续学习视频:python_day6剩余部分和python_day7视频 !一口气学完吧!
续:python单细胞学习笔记-day6
6.2 marker基因可视化
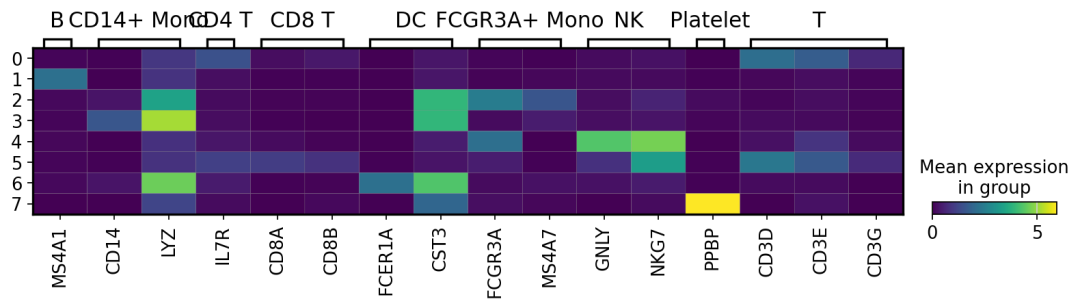
细胞cluster注释,搜集的已知细胞类型的marker基因:
读取进来:
# 读取
df = pd.read_table("markers.txt",header=None)
# 添加列名
df.columns = ['Cell_Type', 'Marker']
# 创建字典,其中每个细胞类型对应一个标记基因列表
markers = df.groupby('Cell_Type')['Marker'].apply(list).to_dict()
markers
# 绘图
sc.pl.dotplot(adata, markers, groupby='leiden',figsize=(15,4),var_group_rotation=0)
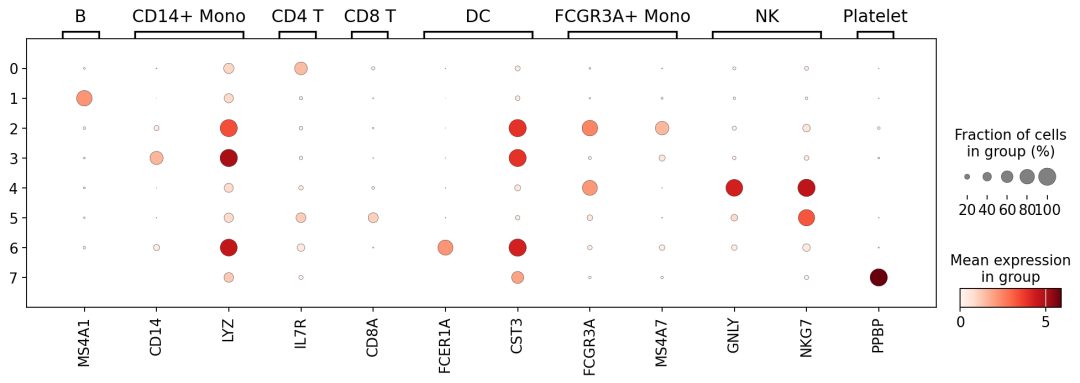
小提琴图:
sc.pl.violin(adata,['PPBP','MS4A1','CD8A'],'leiden')
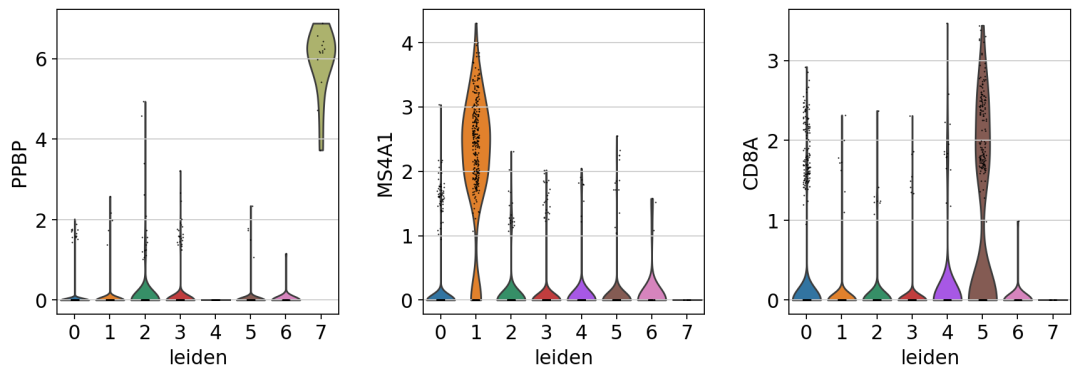
堆叠小提琴图:
sc.pl.stacked_violin(adata, markers, groupby = "leiden",figsize= (16,7),var_group_rotation=0)
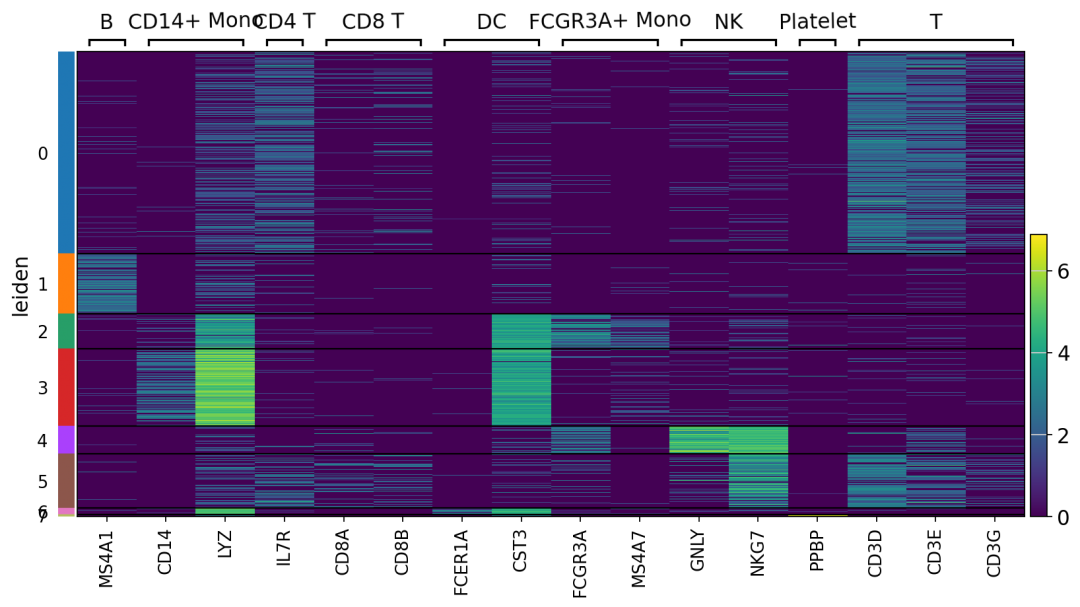
热图:
sc.pl.heatmap(adata, markers, groupby='leiden', var_group_rotation=True,figsize=(12,6))
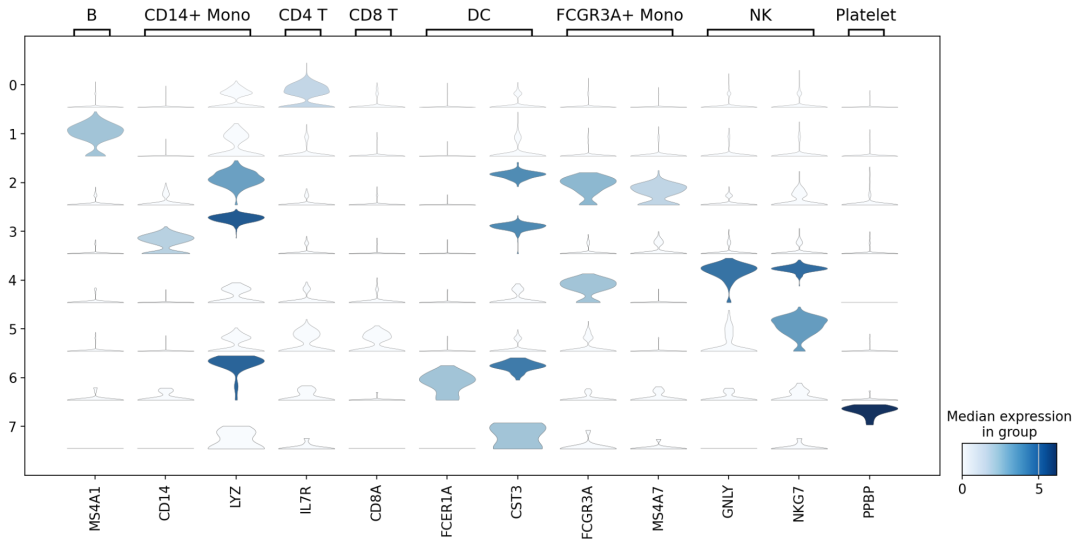
热图2:
sc.pl.matrixplot(adata, markers, groupby='leiden',figsize=(12,2),var_group_rotation= 0)
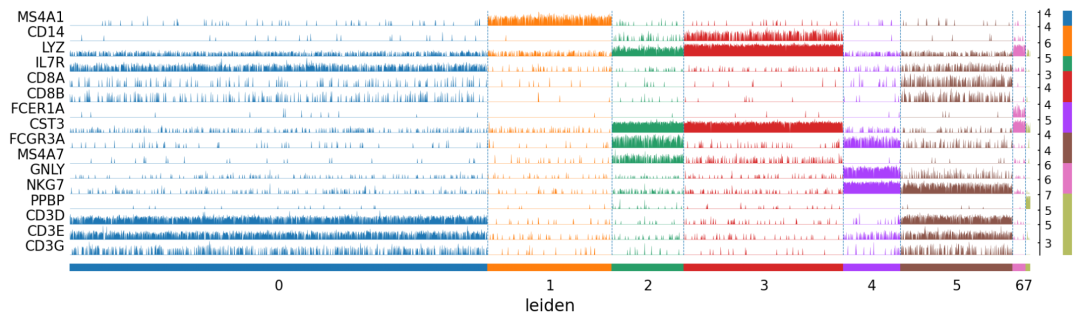
track图:每一条竖线代表一个细胞,线的高度表示基因在细胞中的表达水平
sc.pl.tracksplot(adata,markers,'leiden',figsize=(15,4))
7.细胞类型注释
使用官网的定义:
new_cluster_names = {
'0': "CD4 T",
'1': "B",
'2': "FCGR3A+ Mono",
'3': "NK",
'4': "CD8 T",
'5': "CD14+ Mono",
'6': "DC",
'7': "Platelet"
}
# adata.rename_categories("louvain", new_cluster_names)
adata.obs['leiden'] = adata.obs['leiden'].map(new_cluster_names)
adata.obs['leiden']
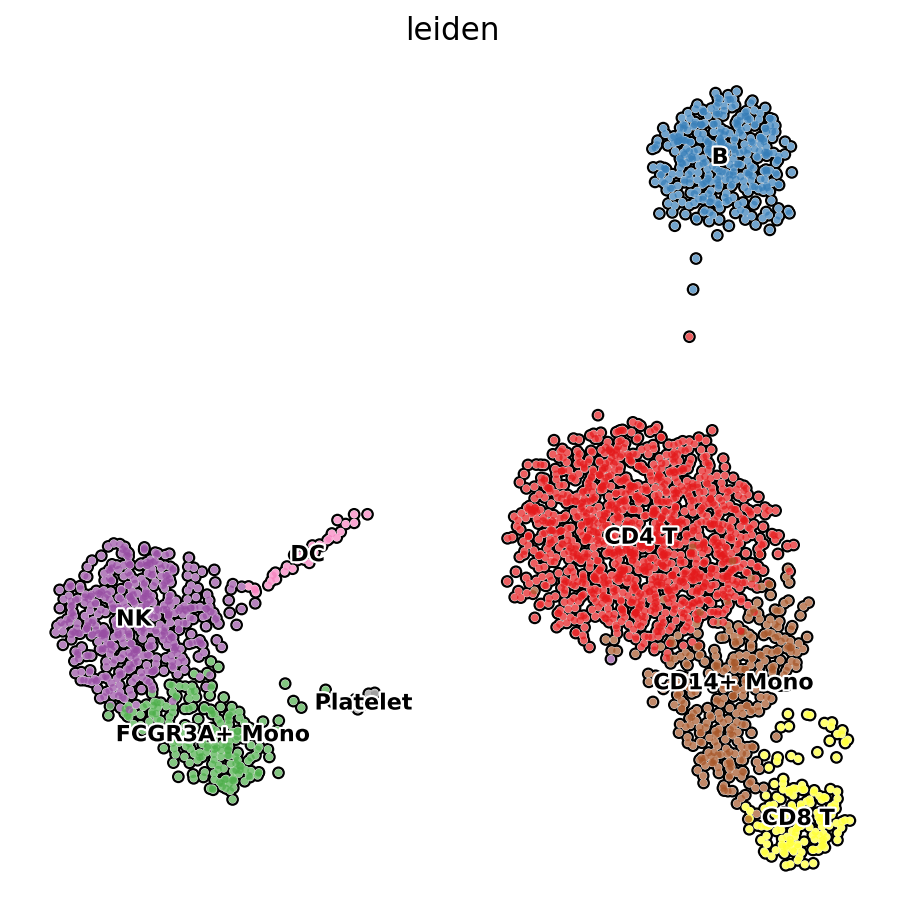
7.1 注释后的umap图
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=(7, 7))
# 使用 scanpy.pl.umap() 绘制 UMAP 图,并将图形绘制到指定的 Axes 对象上
sc.pl.umap(adata, color='leiden', legend_loc='on data',
frameon=False, legend_fontsize=10, legend_fontoutline=2,
add_outline=True, palette="Set1", ax=ax)
# 显示图表
plt.show()
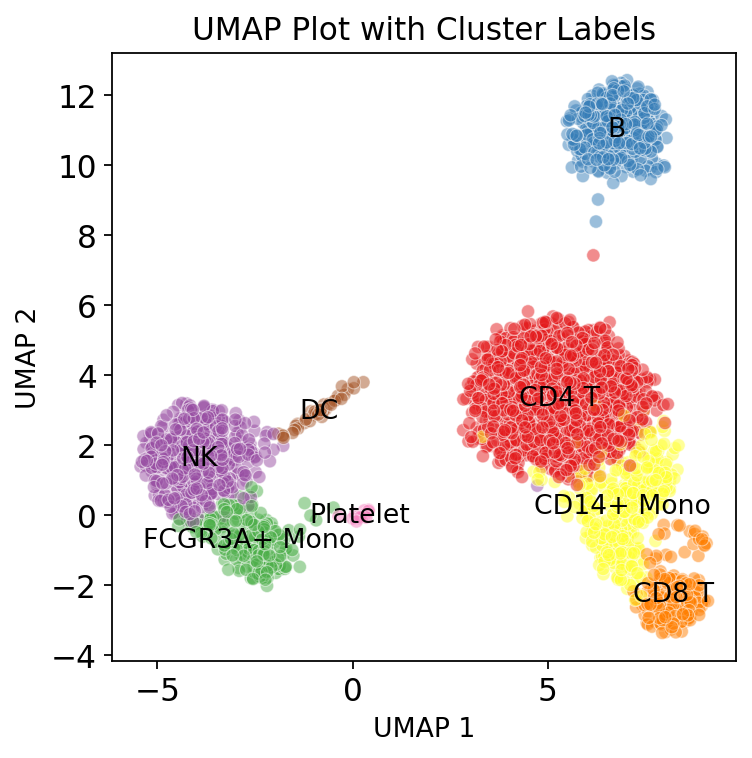
7.2 umap图美化
用seaborn美化umap图:
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
umap_df = pd.DataFrame({
'UMAP_1': adata.obsm['X_umap'][:, 0],
'UMAP_2': adata.obsm['X_umap'][:, 1],
'Cluster': adata.obs['leiden']
})
plt.figure(figsize=(5, 5))
# 使用 seaborn 绘制散点图,按 Cluster 着色
sns.scatterplot(
data=umap_df,
x='UMAP_1',
y='UMAP_2',
hue='Cluster',
palette='Set1',
alpha=0.5,
legend=None
# 不显示默认图例
)
# 计算每个 Cluster 的中心点并添加标签
unique_labels = umap_df['Cluster'].unique()
for label in unique_labels:
label_coords = umap_df[umap_df['Cluster'] == label]
center_x = label_coords['UMAP_1'].mean()
center_y = label_coords['UMAP_2'].mean()
plt.text(center_x, center_y, label, fontsize=12, ha='center', va='center')
# 图片设置
plt.grid(False)
plt.title('UMAP Plot with Cluster Labels', fontsize=14)
plt.xlabel('UMAP 1', fontsize=12)
plt.ylabel('UMAP 2', fontsize=12)
plt.show()
7.3 保存数据
adata.write("pbmc3k_anno.h5ad")
任意分组的差异分析及其可视化
1.加载前面注释好的数据
import scanpy as sc
adata = sc.read_h5ad("pbmc3k_anno.h5ad")
adata
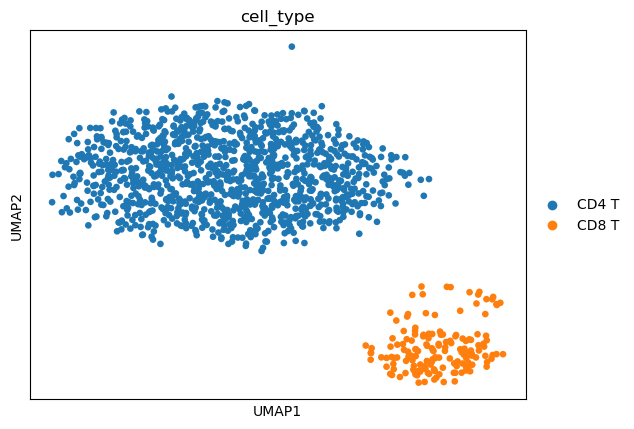
2.提取CD4/CD8 Tcell细胞
adata_subset = adata[adata.obs['leiden'].isin(['CD4 T', 'CD8 T'])].copy()
adata_subset.obs['cell_type'] = adata_subset.obs['leiden'].copy()
sc.pl.umap(adata_subset,color = 'cell_type')
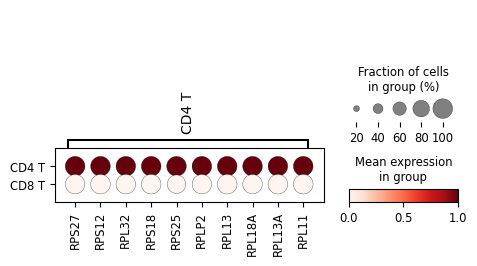
3.找CD4 T细胞和CD8 T细胞之间差异表达的基因
Note:reference是指定对照组
sc.tl.rank_genes_groups(adata_subset, groupby="cell_type", reference="CD8 T",method="wilcoxon",pts=True)
sc.pl.rank_genes_groups_dotplot(
adata_subset, standard_scale="var", n_genes=10
)
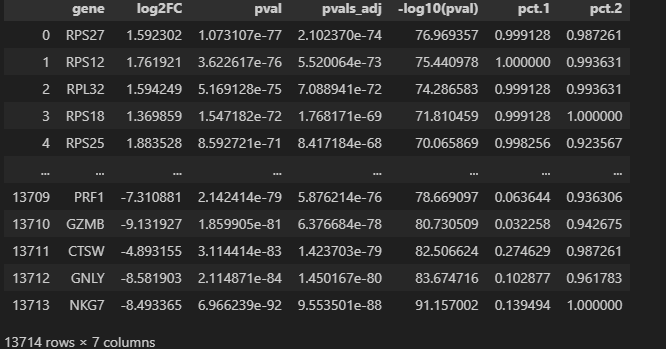
查看得到的差异结果并整理:
# 查看
adata_subset.uns['rank_genes_groups']
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# 提取差异分析结果并构建 DataFrame
df = pd.DataFrame({
'gene': adata_subset.uns['rank_genes_groups']['names']['CD4 T'], # CD4 T 的基因
'log2FC': adata_subset.uns['rank_genes_groups']['logfoldchanges']['CD4 T'], # log2 fold change
'pval': adata_subset.uns['rank_genes_groups']['pvals']['CD4 T'],
'pvals_adj': adata_subset.uns['rank_genes_groups']['pvals_adj']['CD4 T']# p 值
})
# 数据整理
df['-log10(pval)'] = -np.log10(df['pval']) # 计算 -log10(pval)
print(df.head())
# 得到pct
pts = pd.DataFrame({'pct.1': adata_subset.uns['rank_genes_groups']['pts']['CD4 T'],
'pct.2': adata_subset.uns['rank_genes_groups']['pts']['CD8 T']}).reset_index().rename(columns={'index': 'gene'})
pts
# 两者合并
df = df.merge(pts,on = 'gene')
df
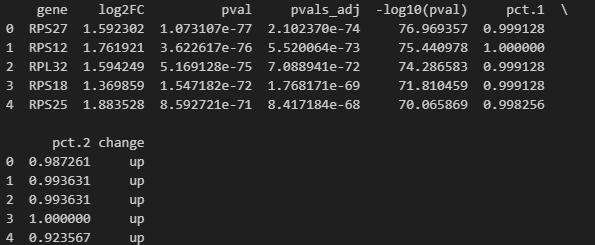
设置阈值挑选上下调:
# 设置阈值
log2fc_threshold = 0.58
pval_threshold = 0.05
k1 = (df['log2FC'] > log2fc_threshold) & (df['pval'] < pval_threshold) #上调
k2 = (df['log2FC'] < -log2fc_threshold) & (df['pval'] < pval_threshold) #下调
k1.value_counts(),k2.value_counts()
# 增加一列上下调
import pandas as pd
df['change'] = df['log2FC'].case_when([
(k1 , "up"),
(k2 , "down"),
(pd.Series(True), "stable")
])
print(df.head())
# 统计差异个数
df.change.value_counts()
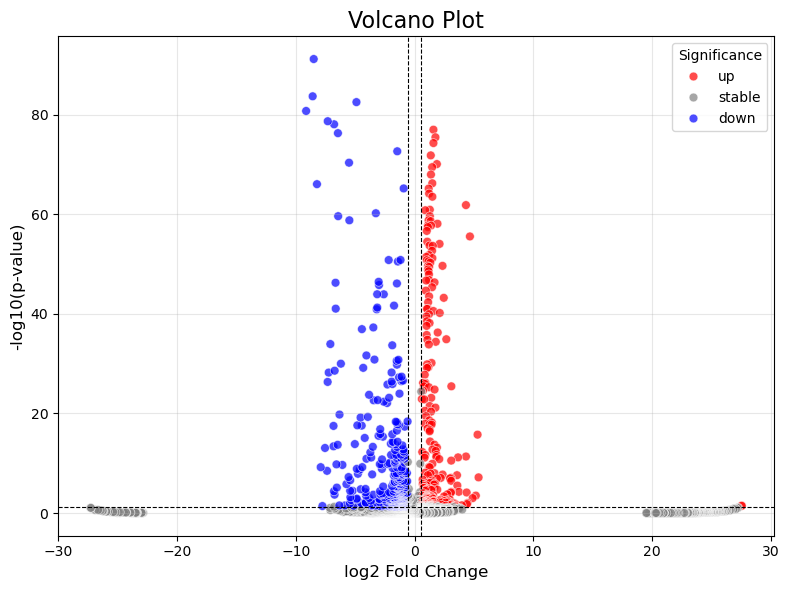
4.绘制火山图
plt.figure(figsize=(8, 6))
sns.scatterplot(
data=df,
x='log2FC',
y='-log10(pval)',
hue='change', # 根据显著性分类上色
palette={
'up': 'red', 'down': 'blue', 'stable': 'grey'},
alpha=0.7,
s=40# 点大小
)
plt.axhline(-np.log10(pval_threshold), color='black', linestyle='--', linewidth=0.8)
plt.axvline(log2fc_threshold, color='black', linestyle='--', linewidth=0.8)
plt.axvline(-log2fc_threshold, color='black', linestyle='--', linewidth=0.8)
plt.title('Volcano Plot', fontsize=16)
plt.xlabel('log2 Fold Change', fontsize=12)
plt.ylabel('-log10(p-value)', fontsize=12)
plt.legend(title='Significance', fontsize=10)
plt.grid(alpha=0.3)
plt.tight_layout()
plt.show()
今天学习到这里~
文末友情宣传